PSE Research Projects
The Expresso project addresses the entire lifecycle of microarray bioinformatics, an area where `computing tools coupled with sophisticated engineering devices [can] facilitate discovery in specialized areas [such as genetics, environment, and drug design]'. Microarrays (sometimes referred to as DNA chips are a relatively new technique in bioinformatics, inspired by miniaturization trends in micro-electronics. Microarray technology is an experimental approach to study all the genes in a given organism simultaneously; it has rapidly emerged as a major tool of investigation in experimental biology. The basic idea is to `print' DNA templates (targets), for all available genes that can be expressed in a given organism, onto a high-density 2D array in a very small area on a solid surface. The goal then is to determine the genes that are expressed when cells are exposed to experimental conditions, such as drought, stress, or toxic chemicals. To accomplish this, RNA molecules (probes) are extracted from the exposed cells and `transcribed' to form complementary DNA (cDNA) molecules. These molecules are then allowed to bind (hybridize) with the targets on the microarray and will adhere only with the locations on the array corresponding to their DNA templates. Typically such cDNA molecules are tagged with fluorescent dyes, so the expression pattern can be readily visualized as an image. Intensity differences in spots will then correspond to differences in expression levels for particular genes. Using this approach, one can `measure transcripts from thousands of genes in a single afternoon'. Microarrays thus constitute an approach of great economic and scientific importance, one whose methodologies are continually evolving to achieve higher value and to fit new uses.
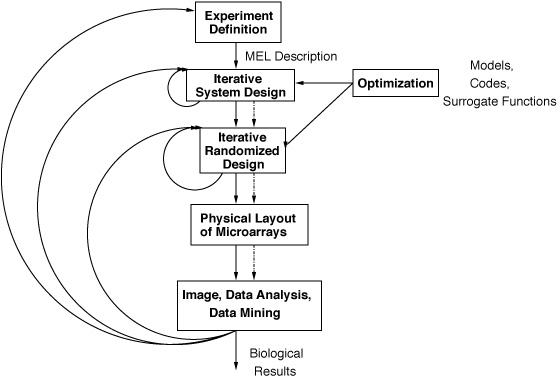
The Expresso PSE is designed to support all microarray activities including experiment design, data acquisition, image processing, statistical analysis, and data mining. Expresso's design incorporates models of biophysical and biochemical processes (to drive experiment management). Sophisticated codes from robotics, physical chemistry, and molecular biology are `pushed' deeper into the computational pipeline. Once designs for experiments are configured, Expresso continually adapts the various stages of a microarray experiment, monitoring their progress, and using runtime information to make recommendations about the continued execution of various stages. Currently, prototypes of the latter three stages of image processing, statistical analysis, and data mining are completely automated and integrated within our implementation.
Expresso's design underscores the importance of modeling both physical and computational flows through a pipeline to aid in biological model refinement and hypothesis generation. It provides for a constantly changing scenario (in terms of data, schema, and the nature of experiments conducted). The ability to provide expressive and high performance access to objects and streams (for experiment management) with minimal overhead (in terms of traditional database functionality such as transaction processing and integrity maintenance) is thus paramount in Expresso.
The design, analysis, and data mining activities in microarray analysis are strongly interactive and iterative. Expresso thus utilizes a lightweight data model to intelligently `close the loop' and address both experiment design and data analysis. The system organizes a database of problem instances and simulations dynamically, and uses data mining to better focus future experimental runs based on results from similar situations. Expresso also uses inductive logic programming (ILP), a relational data mining technique, to model interactions among genes and to evaluate and refine hypothesized gene regulatory networks. One complete instance of the many stages in Expresso has been utilized to study gene expression patterns in Loblolly pine, in a joint project with the Forest Biotechnology group of North Carolina State University.
Last modified: November 5, 2001